Here is for each application a simple procedure to be sure that everything is well installed
Datxplore
- Open Datxplore
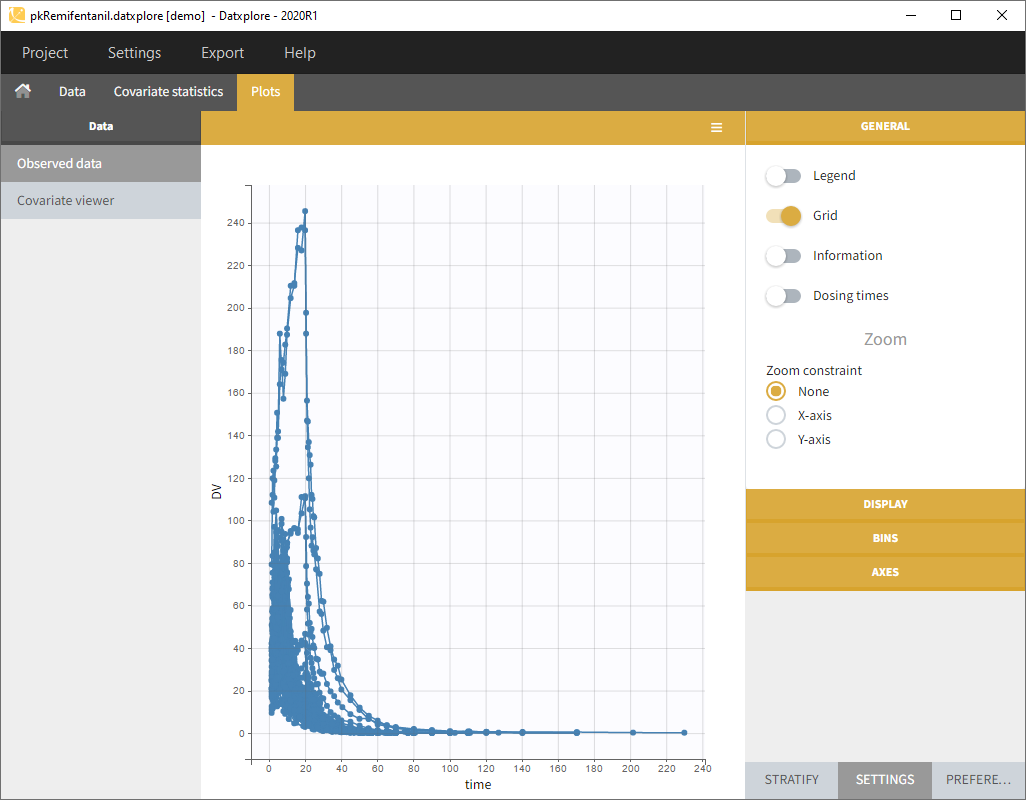
- Go to the Demos (on the left) as on the following figure and load the pkRemifentanil data set
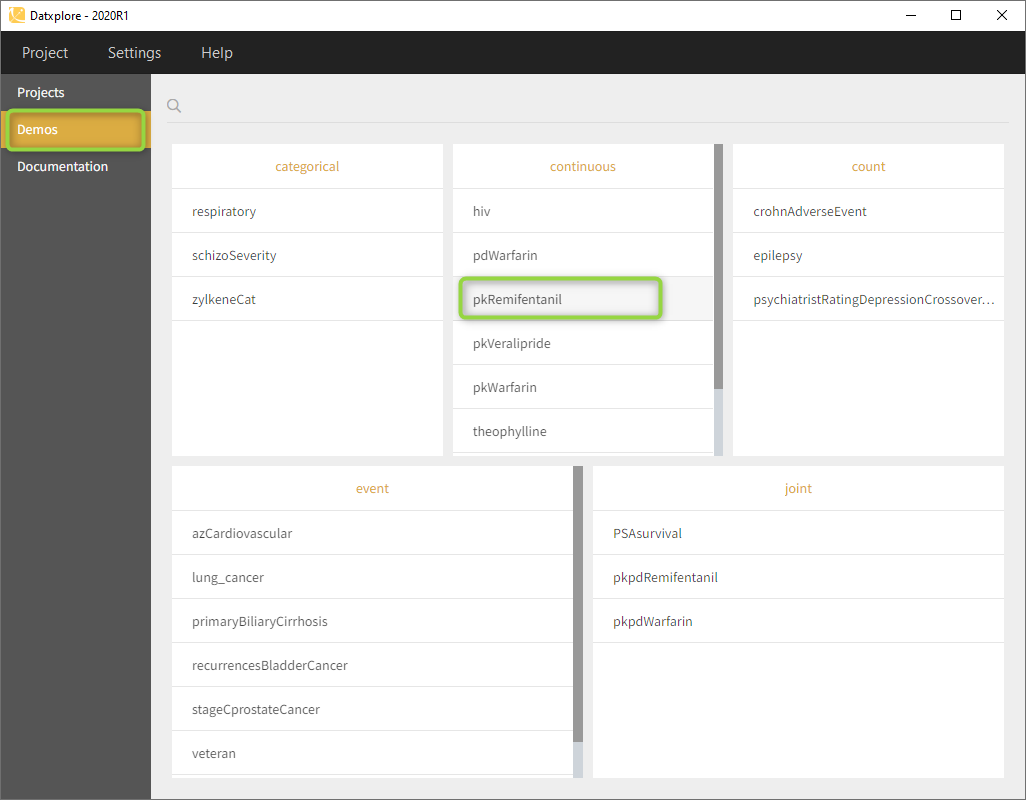
- Datxplore should load the project and display the observed data plot like on the following figure
PKanalix
- Open PKanalix
- Go to the Demos (on the left) and choose project_covariates in 1.basic examples
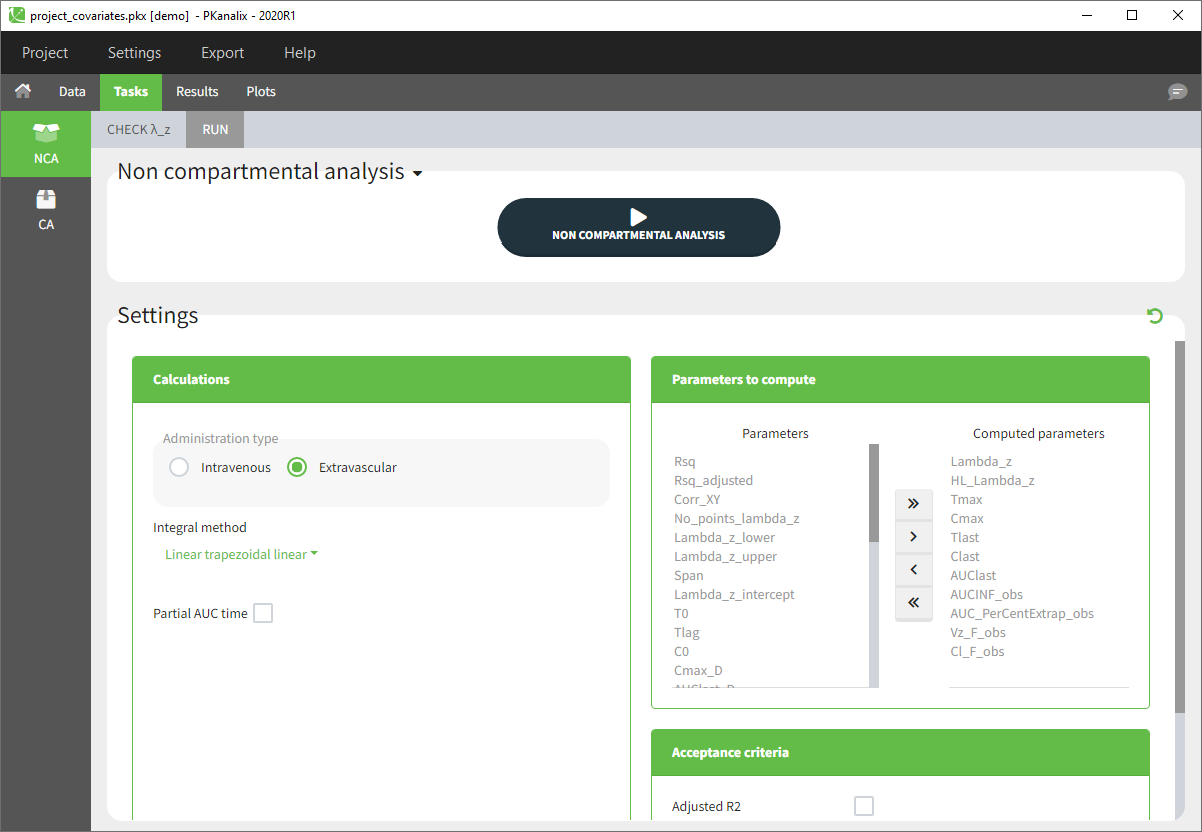
- The Tasks, Results and Plots tabs appear. By default, you are on the Tasks>NCA tab. Click on the button “NON COMPARTMENTAL ANALYSIS” to perform the calculation.
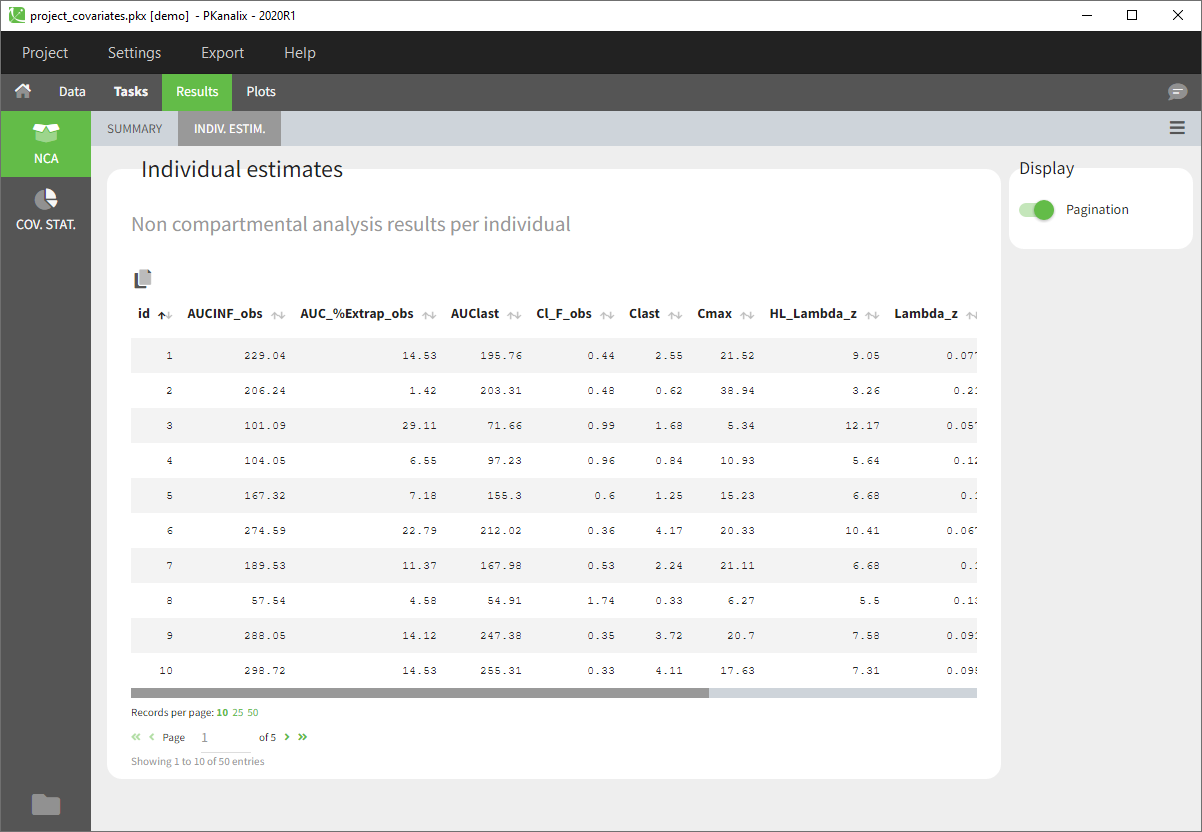
- After the calculation, you are redirected to Results>NCA>Indiv.Estim corresponding to the individual estimates of the NCA parameters.
- You can now click on Plots and by default you’ll see the Individual NCA parameters with respect to the covariates.
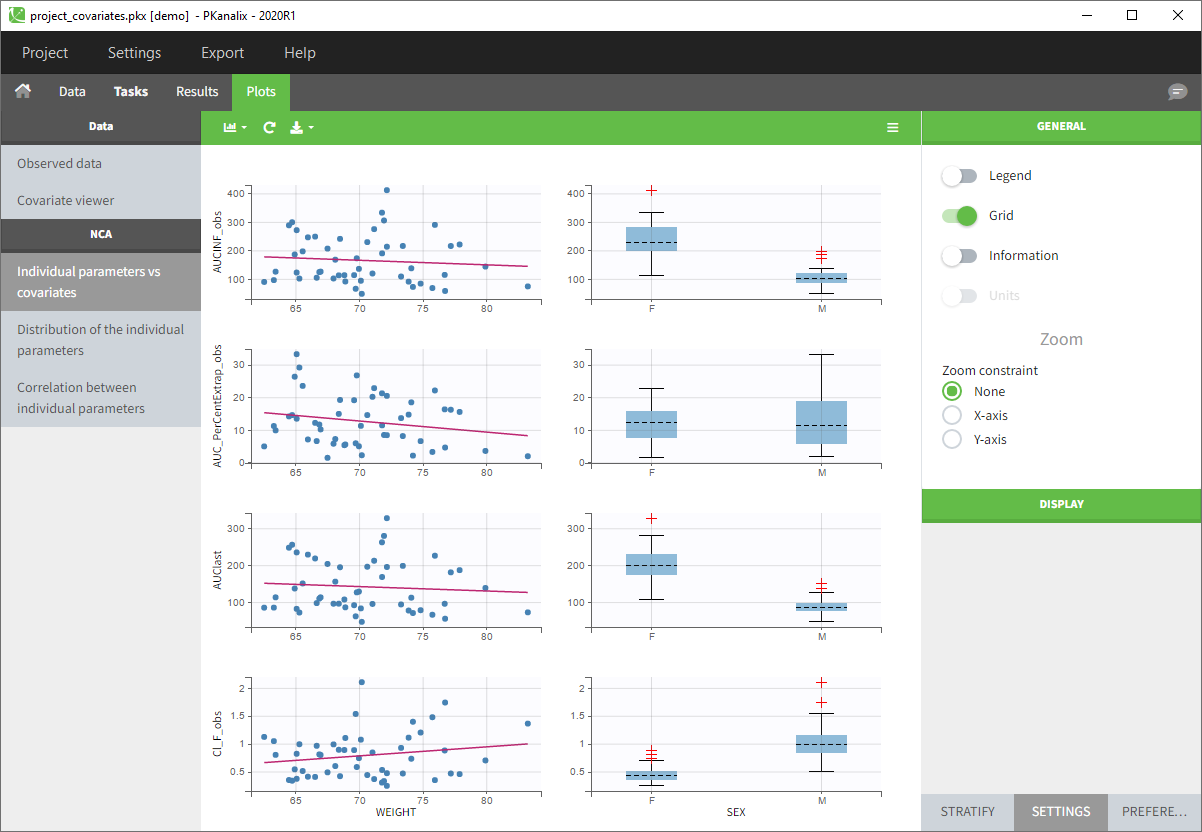
Monolix
- Open Monolix
- Go to the menu Demos and choose theophylline_project in section 1.1 libraries of models

- Monolix loads the project and the interface looks like the following figure
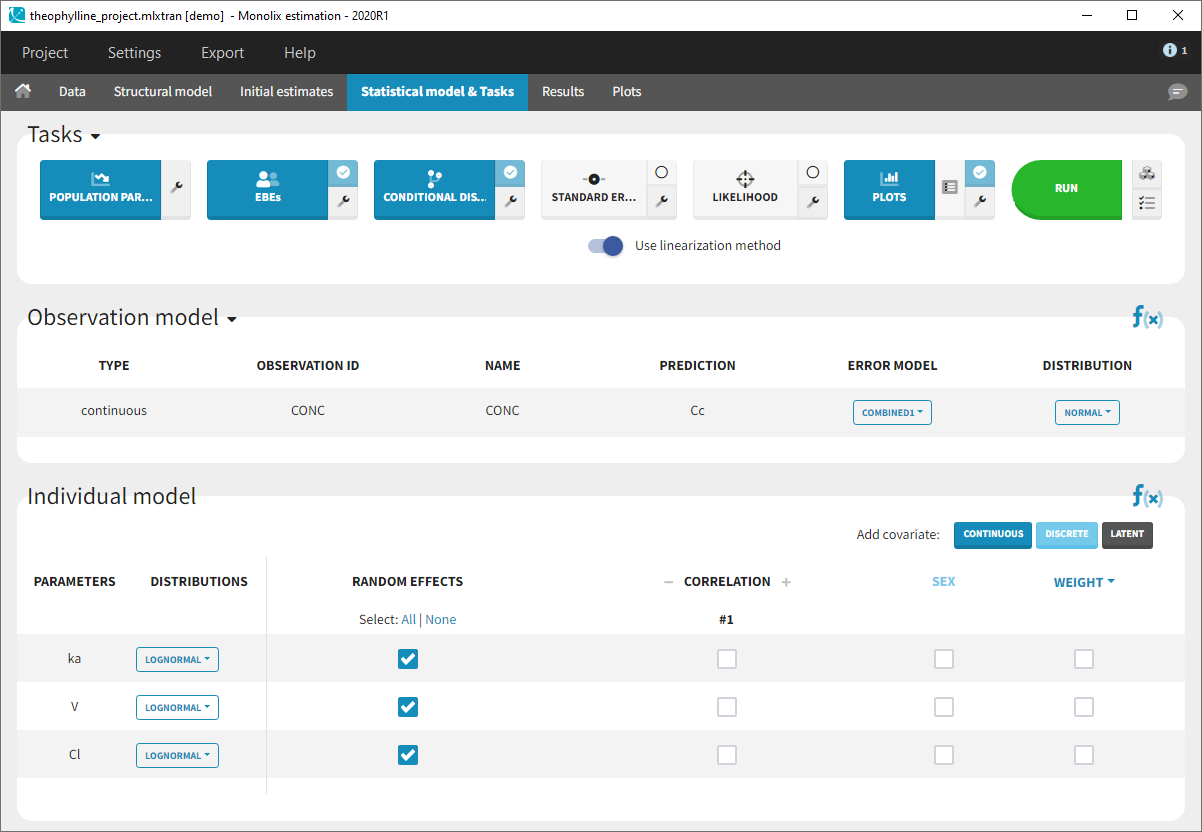
- Clicking on the “Run” button launches the scenario as on the following figure
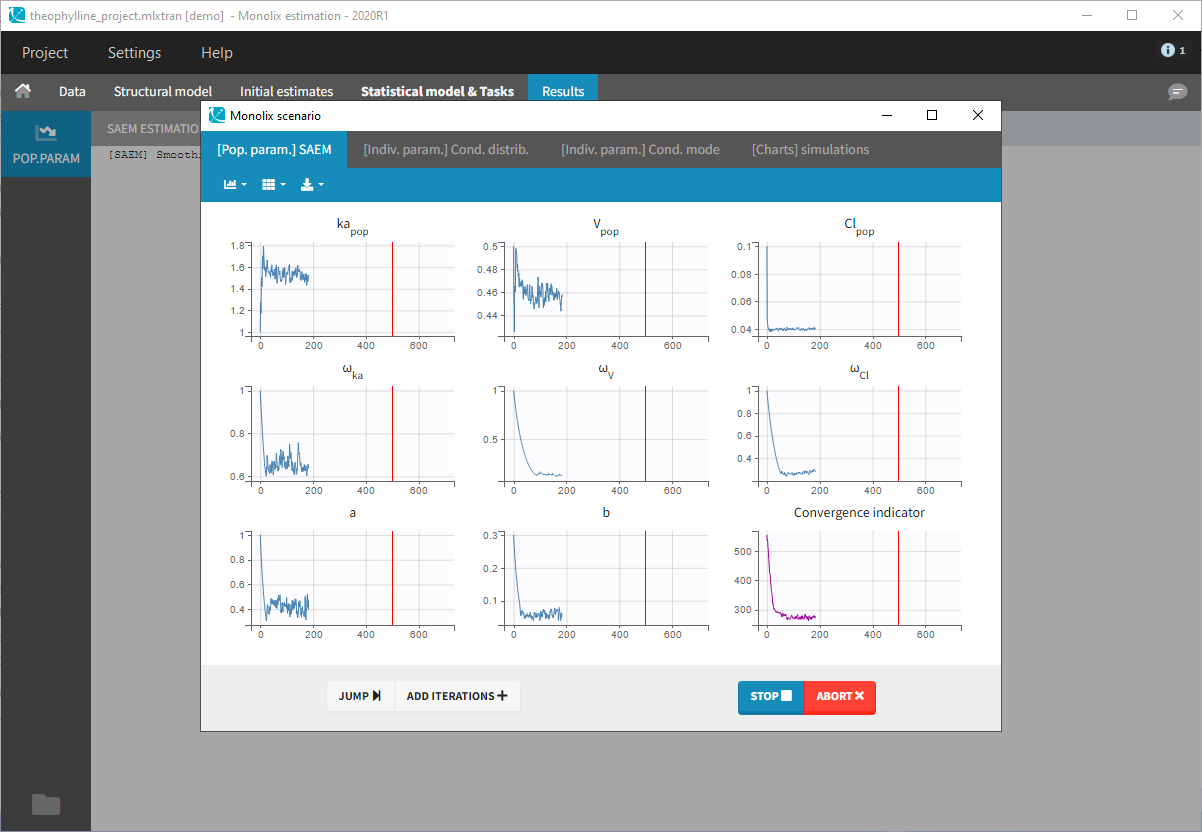
- Close the scenario using the close button and the graphics are displayed behind on the Individual fits plot
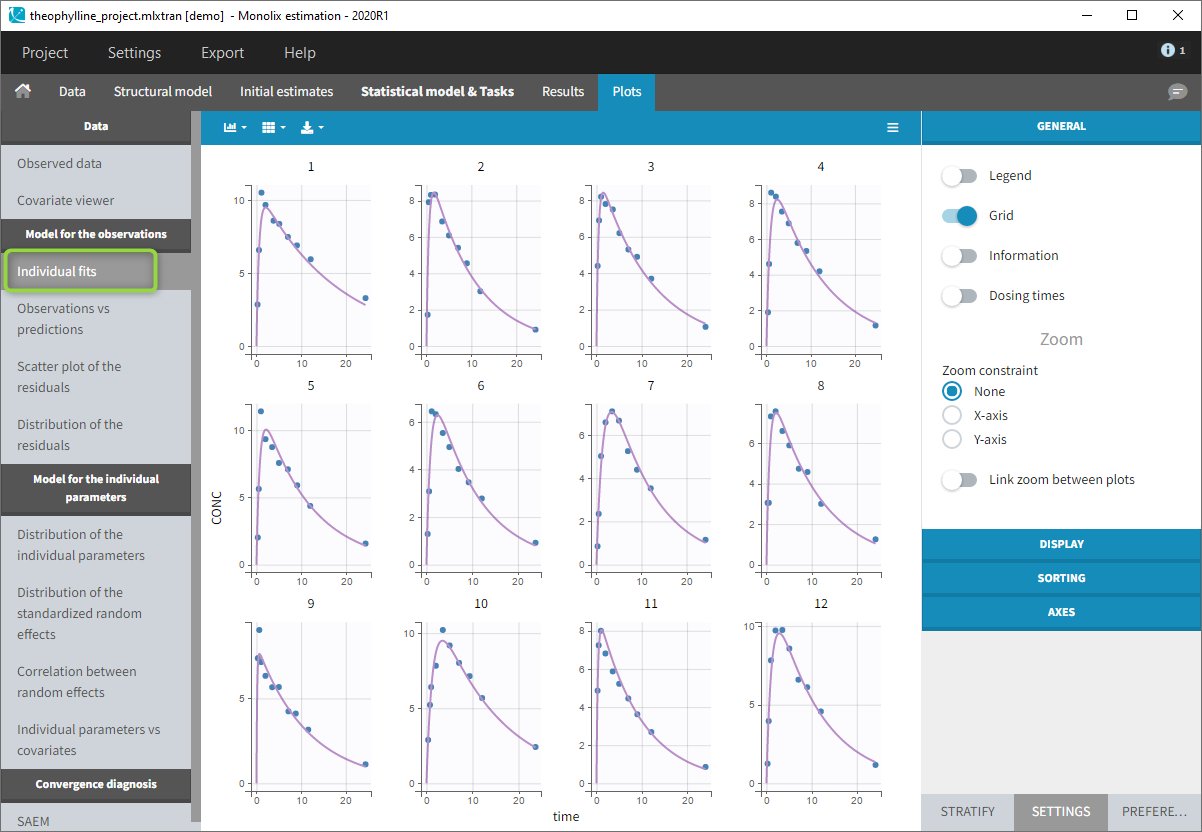
Simulx
- Open Simulx
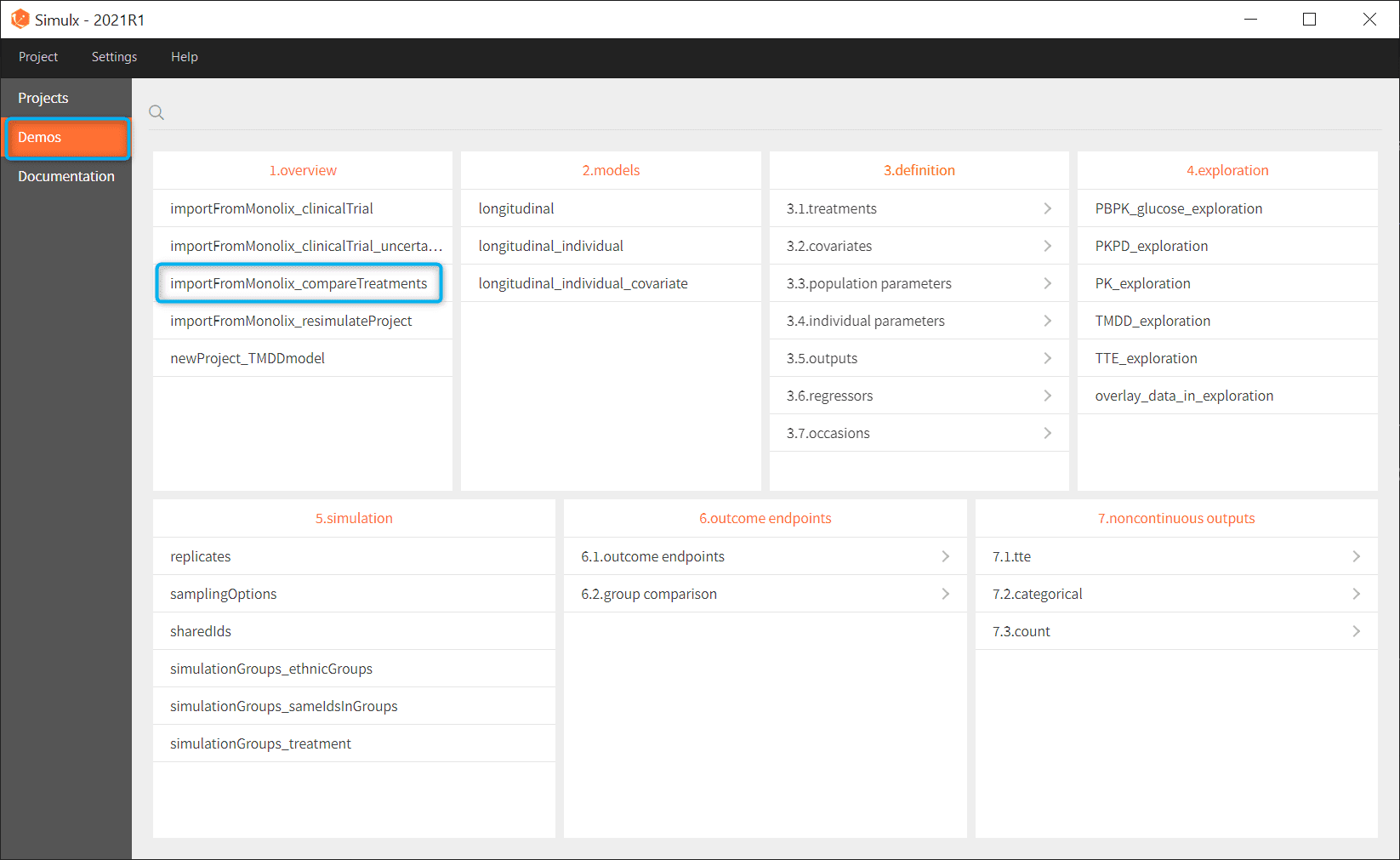
- Go to the menu Demos and choose importFromMonnolix_compareTreatments in section 1.overview
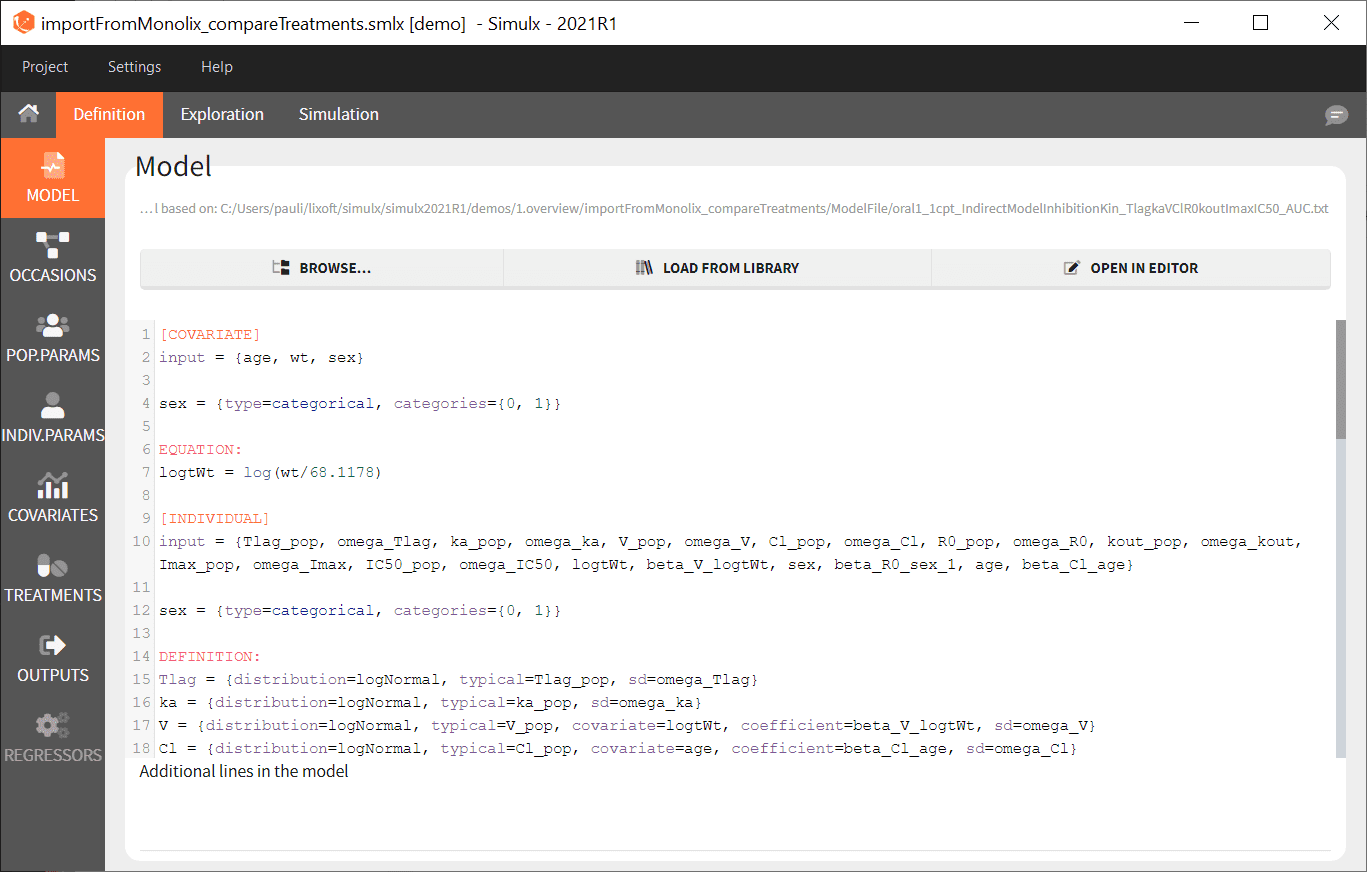
- Simulx loads the project and the interface looks like the following figure
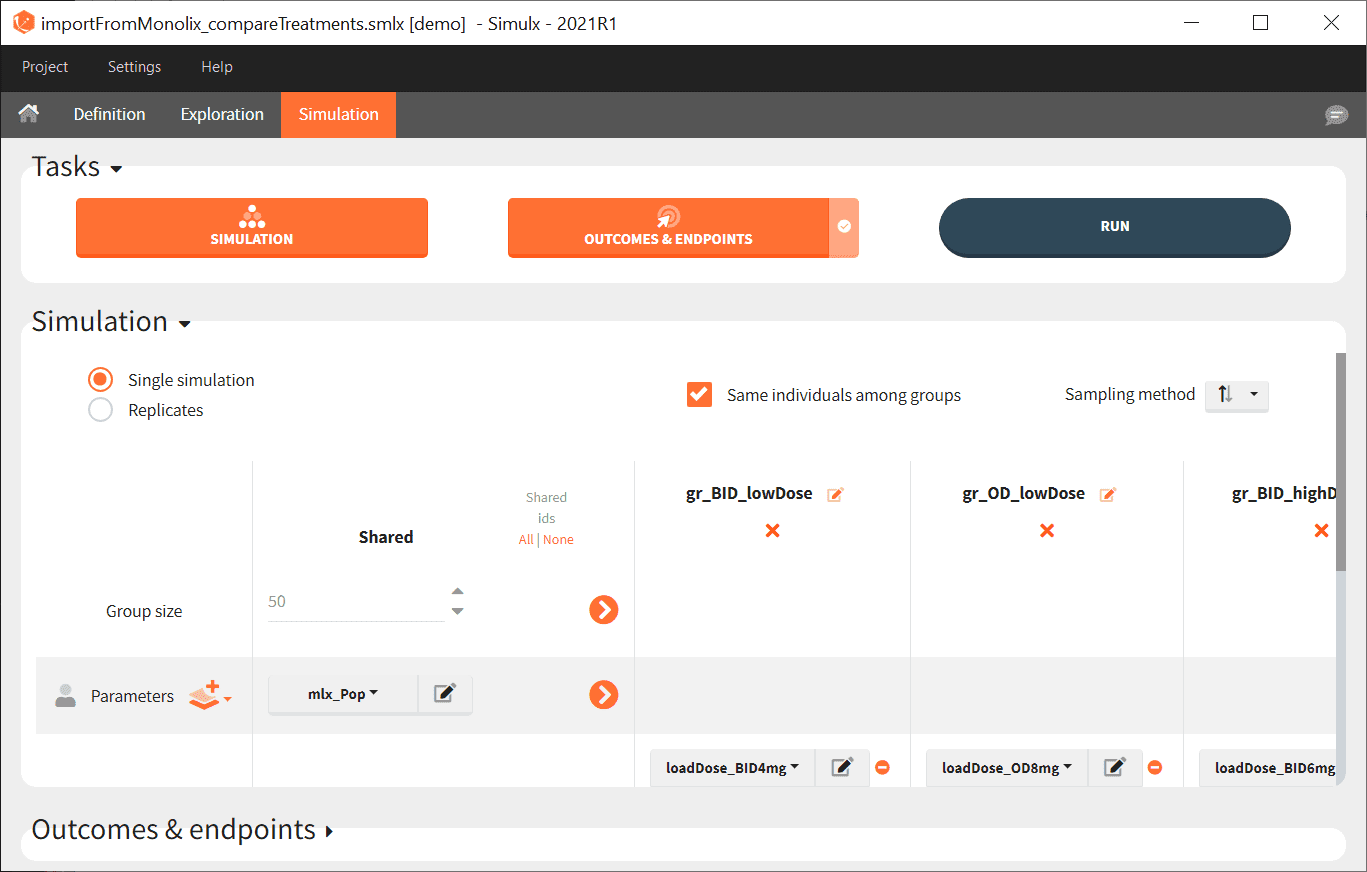
- Go on the Simulation tab where the simulation is defined as shown here and click on RUN
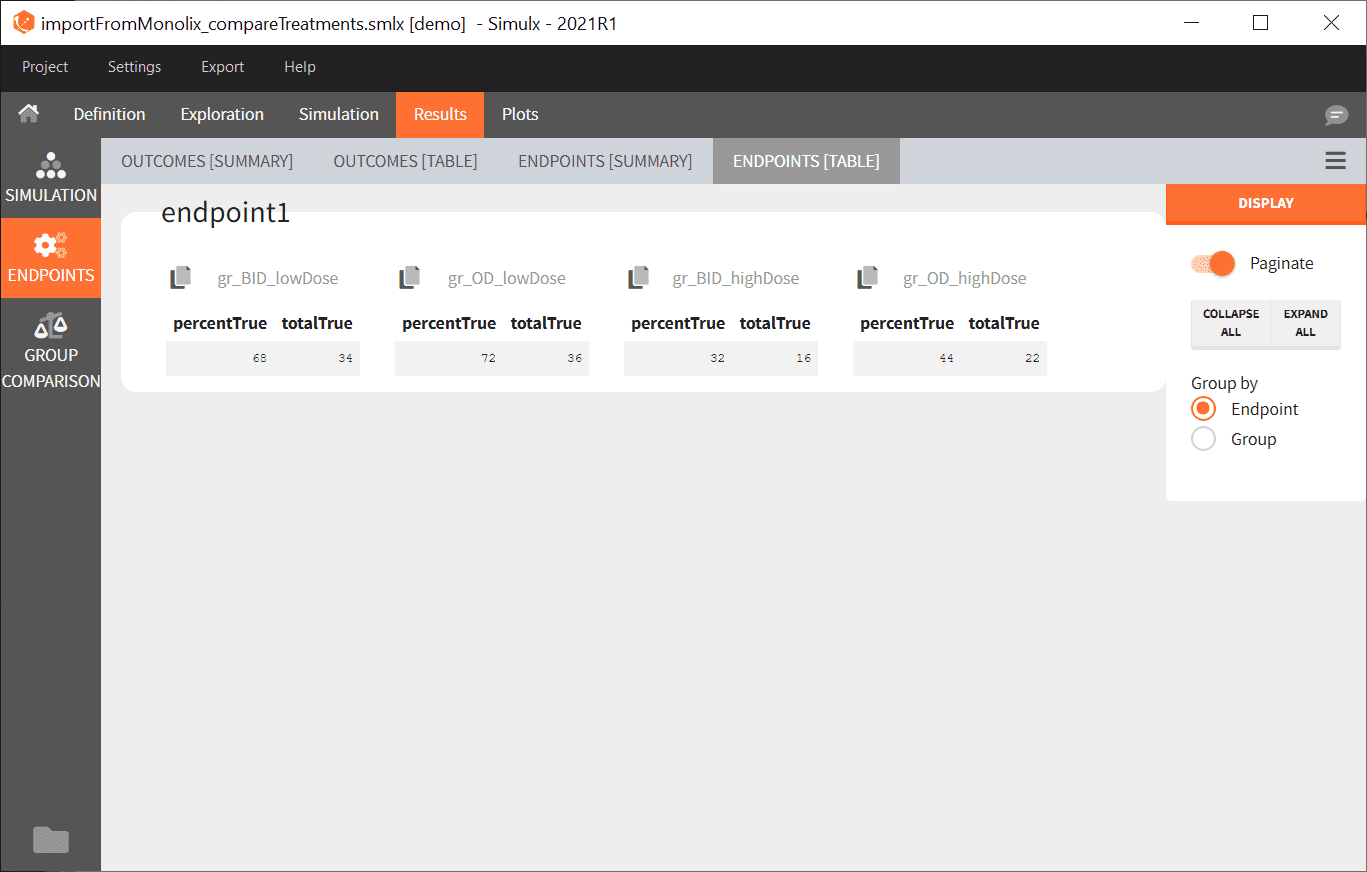
- The numerical results of the simulations are the displayed
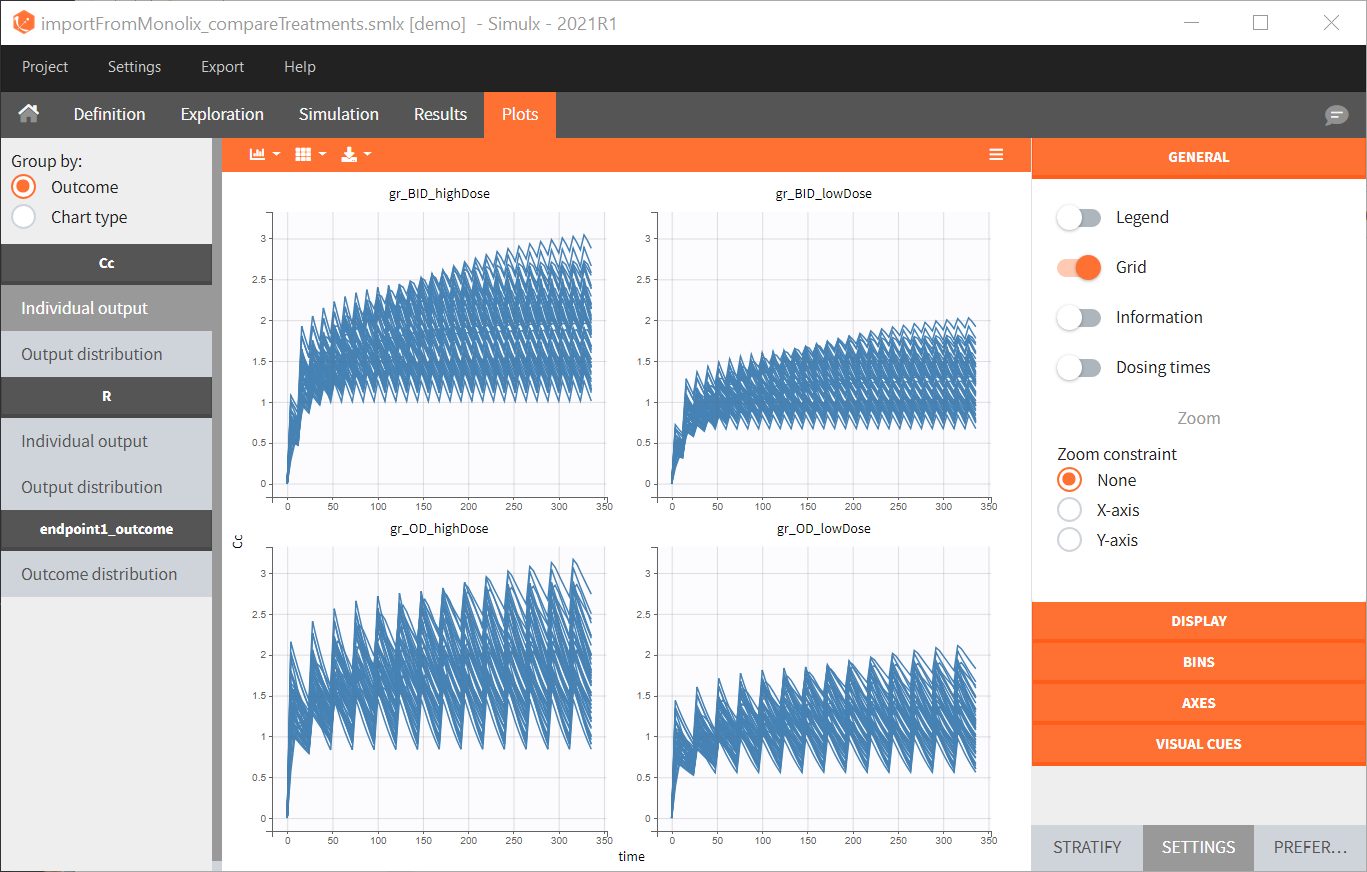
- By clicking on the Plots tab, you can see the numerical results as here.